UMAP on RAPIDS (15x Speedup)
Exploring the new GPU acceleration library from NVIDIA
Mar 30, 2019 · 457 words · 3 minute read

RAPIDS
RAPIDS is a collection of Python libraries from NVIDIA that enables the users to do their data science pipelines entirely on GPUs. The two main components are cuDF and cuML. The cuDF library provides Pandas-like data frames, and cuML mimics scikit-learn. There’s also a cuGRAPH graph analytics library that have been introduced in the latest release (0.6 on March 28).
The RAPIDS suite of open source software libraries gives you the freedom to execute end-to-end data science and analytics pipelines entirely on GPUs. RAPIDS is incubated by NVIDIA® based on years of accelerated data science experience. RAPIDS relies on NVIDIA CUDA® primitives for low-level compute optimization, and exposes GPU parallelism and high-bandwidth memory speed through user-friendly Python interfaces.
This NeurIPS 2018 talk provides an overview and vision of RAPIDS when it was launched:
The RAPIDS project also provides some example notebook. Most prominently the cuDF + Dask-XGBoost Mortgage example. (The link to the dataset in the notebook is broken. Here’s the correct one.)
Does RAPIDS Help in Smaller Scales?
The mortgage datasets are huge. The 17-year one takes 195 GB, and the smallest 1-year one takes 3.9 GB. The parallel processing advantage of GPUs is obvious for larger datasets. I wonder if smaller datasets can benefit from RAPIDS as well?
In the following example, we use the Fashion-MNIST dataset (~30MB) and run UMAP (Uniform Manifold Approximation and Projection) algorithm for dimension reduction and visualization. We compare the performances of the CPU implementation and the RAPIDS implementation.
It turns out the RAPIDS implementation can be 15x times faster (caveat: I only ran the experiments two times, but they returned roughly the same results):
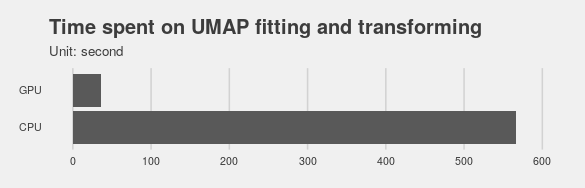
The visualization shows the end products from CPU and RAPIDS implementation are very similar.
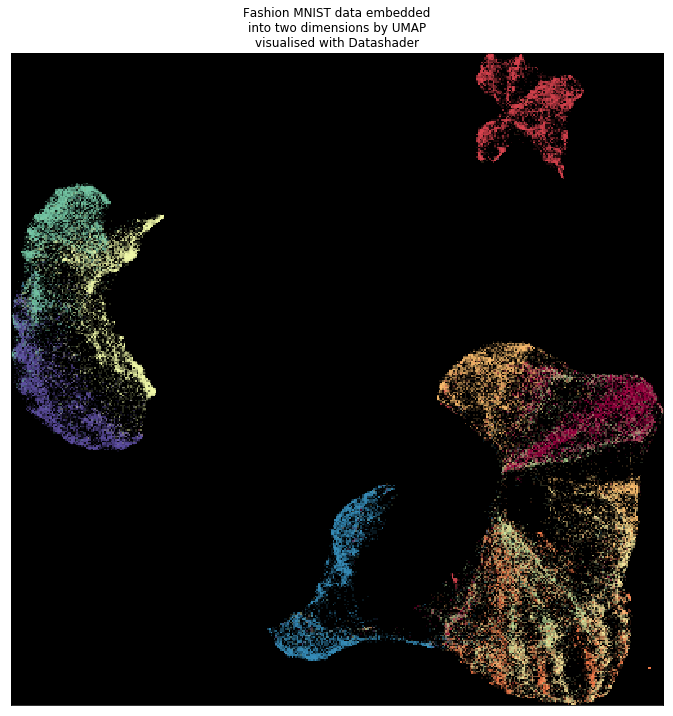
From the CPU implementation.
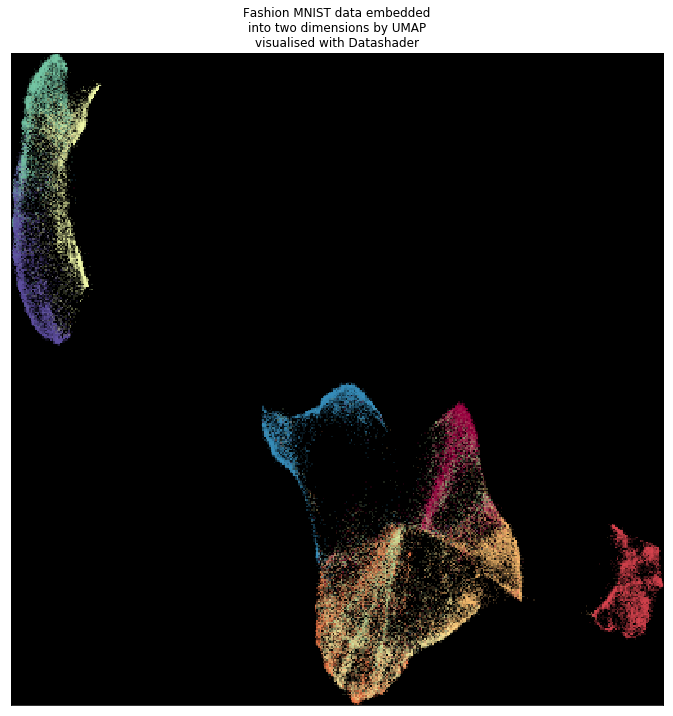
From the RAPIDS implementation.
The RAPIDS implementation lacks some features, though. For example, the random state cannot be set, and the distance metric is fixed (it is not stated in the documentation. I assume it is Euclidean.). But if you don’t need those features, RAPIDS can save you a lot of time.
Source Code and Environment Setup
I used the official Docker image (rapidsai/rapidsai:0.6-cuda10.0-runtime-ubuntu18.04-gcc7-py3.7) to run the following notebook.
To pull the image and start an container, run:
$ docker pull `rapidsai/rapidsai:0.6-cuda10.0-runtime-ubuntu18.04-gcc7-py3.7`
$ docker run --runtime=nvidia \
--rm -it \
-p 8888:8888 \
-p 8787:8787 \
-p 8786:8786 \
`rapidsai/rapidsai:0.6-cuda10.0-runtime-ubuntu18.04-gcc7-py3.7`
(rapids) root@container:/rapids/notebooks# bash utils/start-jupyter.sh`
Fin
Thanks for reading! This is a short post introducing RAPIDS and presents a simple experiments showing how RAPIDS can help you speed up the UMAP algorithm. RAPIDS is still a very young project, and very new to me. I’ll try to use RAPIDS in my future data analytics projects and maybe write more posts about it. Stay tuned!